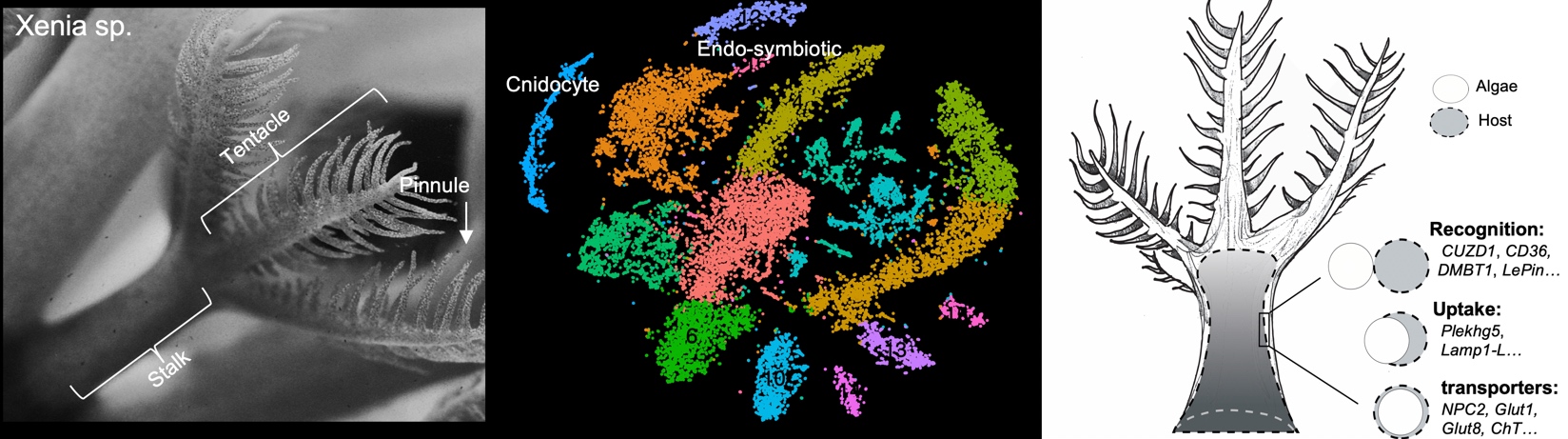
This repository contains the code and analyses associated with a single-cell RNA-seq study of Xenia sp. presented in the following manuscript:
Minjie Hu, Xiaobin Zheng, Chen-Ming Fan, and Yixian Zheng (2020) Lineage dynamics of the endosymbiotic cell type in the soft coral Xenia https://www.nature.com/articles/s41586-020-2385-7
The transcriptome and the genome can be accessed via Carnegie Coral & Marine Organisms
Raw data used for de novo genome assembly, gene prediction and scRNA-seq are accessible at SRA (Bioproject: PRJNA548325)
Selected/final R analysis objects are available from Carnegie Coral & Marine Organisms
R Markdown documents with analysis code (also available as knitted html files).
1_CreateSeuratObject.Rmd
- Filter data and create Seurat object for each scRNA library
2_ClusterAnalysis.Rmd
- Integrate data from non-regeneration samples, initial clustering, and determine markers for each cluster.
3_SymbioticCellIdentification.Rmd
- Compare with FACS transcriptome and identify endosymbiotic cell cluster
4_RNAvelocityAnalysis.Rmd
- RNA velocity analysis for regeneration sample, defining early and late endosymbotic cell
5_MonoclePseudotimeAnalysis.Rmd
- Monocle pseudotime analysis for all endo-symbiotic cells, defining pre-endosymbiotic, transition1, mature, transition2, and post-endosymbiotic states.
All data files needed to repeat the analysis can be fetched through following commands:
git clone https://github.com/ciwemb/endosymbiosis
wget -r -np -nH --reject="index.html*" \
http://cmo.carnegiescience.edu/endosymbiosis/