-
Notifications
You must be signed in to change notification settings - Fork 162
nanocube ready dmp
Suppose you have some application that has nothing to do with
geographical maps, latitudes, and longitudes, but you still have
data with spatial dimensions (e.g. floor plan of a building), categorical
dimensions and a temporal dimension. You can also use nanocubes to store
and query your data. The way to do that is to learn how to generate
.dmp files that are nanocube-ready and bypass the binning
tools that come with the nanocube distribution (i.e. old ncdmp,
nanocube-binning-dmp, nanocube-binning-csv). To model spatial
domains and feed it's data into nanocube the way to do it is to model
it as a square grid with 2^n x 2^n cells and use a quadtree dimension
to store it. Here is an example
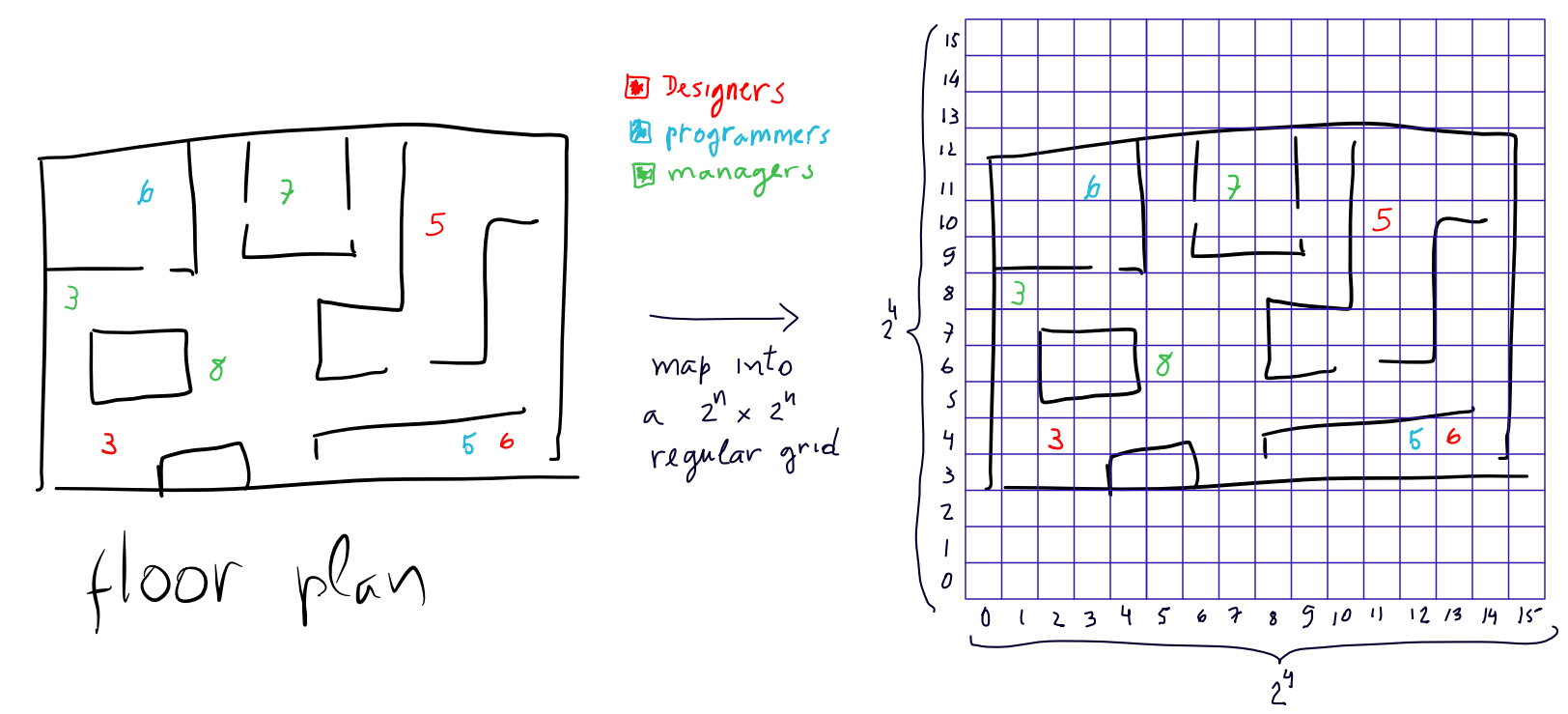
In this example let's assume the employee map in our floorplan (designers, programmers and managers) is for time instant 1. So the data we want to feed into the nanocube is the following:
location job time count
2,3 designer 1 3
1,8 manager 1 3
3,11 programmer 1 6
5,6 manager 1 8
11,10 designer 1 5
12,4 programmer 1 5
13,4 designer 1 6
A nanocube ready .dmp file for this table is the following
name: designer
field: location nc_dim_quadtree_4
field: job nc_dim_cat_1
field: time nc_dim_time_2
field: count nc_var_uint_4
valname: job 0 designer
valname: job 1 manager
valname: job 2 programmer
<records-encoded-in-binary>
The binary encoding of a dimension of type nc_dim_quadtree_<levels>
expects two four bytes integers: one for the x grid location and
another one for the y grid location (coordinates in a 2^n-1 x 2^n-1
grid). The binary encoding for nc_dim_cat_<b_bytes> expects an
integer with b byte for the category (the mapping to actual names is
given on the valname lines). The nc_dim_time_<t_bytes> expects an
integer with t bytes as the timestamp. Finally the
nc_var_uint_<c_bytes> indicates how many units we want to add in
that (location, job, time) "bin". Everything shouls be encoded in
little endian. So each row from our table should map to a 4 + 4 + 1 + 2 + 4 = 15 bytes record. You can investigate the outcome of
nanocube-binning-dmp and nanocube-binning-csv to get more examples
of nanocube ready .dmp files.
Once a file like the described above is generated we can feed it directly into a nanocube process.
cat floorplan-nanocube-ready.dmp | nanocube-leaf -q 29512
importante: you need a specific binary to make this example work that is not built by default by the standard nanocube distribution. See section below on compiling a non-standard nanocube binary.
Here is the nanocube .dmp file for this example floorplan-nanocube-ready.dmp:
With the nanocube above running, here are some queries we can run:
# total number of employees in my map
http://localhost:29512/count
{ "layers":[ ], "root":{ "val":36 } }
# total number of programmers
http://localhost:29512/count.r("job",1)
{ "layers":[ ], "root":{ "val":11 } }
# total number of employees per job
http://localhost:29512/count.a("job",dive([],1))
{ "layers":[ "anchor:job" ], "root":{ "children":[ { "path":[0], "val":14 }, { "path":[1], "val":11 }, { "path":[2], "val":11 } ] } }
# total number of employees in the bottom-left quadrant
http://localhost:29512/count.r("location",[0])
{ "layers":[ ], "root":{ "val":11 } }
# total number of employees per quadrant
http://localhost:29512/count.a("location",dive([],1))
{ "layers":[ "anchor:location" ], "root":{ "children":[ { "path":[3], "val":5 }, { "path":[2], "val":9 }, { "path":[1], "val":11 }, { "path":[0], "val":11 } ] } }
# total number of employees per quadrant as img (2x2)
http://localhost:29512/count.a("location",dive([],1),"img")
{ "layers":[ "anchor:location" ], "root":{ "children":[ { "x":1, "y":1, "val":5 }, { "x":0, "y":1, "val":9 }, { "x":1, "y":0, "val":11 }, { "x":0, "y":0, "val":11 } ] } }
# total number of employees per quadrant as img (16x16)
http://localhost:29512/count.a("location",dive([],4),"img")
{ "layers":[ "anchor:location" ], "root":{ "children":[ { "x":11, "y":10, "val":5 }, { "x":3, "y":11, "val":6 }, { "x":1, "y":8, "val":3 }, { "x":12, "y":4, "val":5 }, { "x":13, "y":4, "val":6 }, { "x":2, "y":3, "val":3 }, { "x":5, "y":6, "val":8 } ] } }
# total number of employees per quadrant as img (16x16) and job
http://localhost:29512/count.a("location",dive([],4),"img").a("job",dive([],1))
{ "layers":[ "anchor:location", "anchor:job" ], "root":{ "children":[ { "x":11, "y":10, "children":[ { "path":[0], "val":5 } ] }, { "x":3, "y":11, "children":[ { "path":[1], "val":6 } ] }, { "x":1, "y":8, "children":[ { "path":[2], "val":3 } ] }, { "x":12, "y":4, "children":[ { "path":[1], "val":5 } ] }, { "x":13, "y":4, "children":[ { "path":[0], "val":6 } ] }, { "x":2, "y":3, "children":[ { "path":[0], "val":3 } ] }, { "x":5, "y":6, "children":[ { "path":[2], "val":8 } ] } ] } }
Here is a python script to generate a nanocube ready .dmp file for
the floorplan example python script:
import sys
import struct
data='''
location job time count
2,3 designer 1 3
1,8 manager 1 3
3,11 programmer 1 6
5,6 manager 1 8
11,10 designer 1 5
12,4 programmer 1 5
13,4 designer 1 6
'''
job_code = { 'designer':0, 'programmer':1, 'manager':2 }
columns = None
class Record(object):
def __init__(self):
pass
records = []
# assuming there are dimensions called time, location, count.
# other dimensions will be considered categorical dimensions
# of up to 255 different values
LOCATION, TIME, COUNT, CATEGORICAL = range(4)
def type_from_name(c):
if c == 'location':
return LOCATION
elif c == 'time':
return TIME
elif c == 'count':
return COUNT
else:
return CATEGORICAL
# collect records
line_no = 0
for line in data.split('\n'):
line_no += 1
line = line.strip()
if len(line) == 0:
continue
print line
tokens = line.split()
print tokens
# read header
if columns == None:
columns = [(x,type_from_name(x)) for x in tokens if len(x) > 0]
else:
tokens = [x for x in tokens if len(x) > 0]
if len(tokens) != len(columns):
raise Exception("Problem on line %d: tokens and columns are different")
record = Record()
for i in xrange(len(tokens)):
column_name, column_type = columns[i]
value = tokens[i]
if column_type == LOCATION: # x,y
value = [int(x) for x in value.split(',')]
elif column_type == TIME or column_type == COUNT:
value = int(value)
elif column_type == CATEGORICAL:
codes = eval("%s_code" % column_name) # assuming there is a map with the coding of that category
# assume categorical
value = codes[value]
record.__dict__[column_name] = value
records.append(record)
print record.__dict__
# write nanocube-ready .dmp file
ostream = open('floorplan-nanocube-ready.dmp','w')
def field_type_for(column_type):
grid_levels = 4 # grid of 2^4 to 2^4
if column_type == LOCATION:
return "nc_dim_quadtree_" + str(grid_levels)
elif column_type == TIME:
return "nc_dim_time_2" # make it 2 bytes time bins can go from 0 to 2^16-1
elif column_type == COUNT:
return "nc_var_uint_4" # make it 4 bytes count bins can go from 0 to 2^32-1
elif column_type == CATEGORICAL:
return "nc_dim_cat_1"
ostream.write("name: floorplan\n")
for i in xrange(len(columns)):
column_name, column_type = columns[i]
ostream.write("field: %s %s\n" % (column_name, field_type_for(column_type)))
for i in xrange(len(columns)):
column_name, column_type = columns[i]
if column_type == CATEGORICAL:
codes = eval("%s_code" % column_name) # assuming there is a map with the coding of that category
inverted_codes = dict([(v,k) for k,v in codes.iteritems()])
keys = sorted(inverted_codes.keys())
for k in keys:
ostream.write("valname: %s %d %s\n" % (column_name, k, inverted_codes[k]))
ostream.write("\n") # end of header indication
for r in records: # write binary records
for column_name,column_type in columns:
if column_type == LOCATION:
ostream.write(struct.pack("<II",r.location[0],r.location[1]))
elif column_type == TIME:
ostream.write(struct.pack("<H",r.time))
elif column_type == COUNT:
ostream.write(struct.pack("<I",r.count))
elif column_type == CATEGORICAL:
ostream.write(struct.pack("<B",r.__dict__[column_name]))At its current state (release 3.1), we have to have a specific
nanocube binary for each permutation of field types. For example,
in the foorplan case above we need a binary called: nc_q4_c1_u2_u4.
You can custom binaries by editing the file src/Makefile.am and
add to the list bin_PROGRAMS the binary you need:
bin_PROGRAMS = \
nanocube-binning-dmp \
nanocube-leaf \
nc_q25_u2_u4 \
nc_q25_c1_u2_u8 \
nc_q25_c1_u2_u4 \
nc_q25_c1_c1_u2_u4 \
nc_q25_c1_c1_c1_u2_u4 \
nc_q25_q25_u2_u4 \
nc_q25_c2_u2_u4 \
nc_q4_c1_u2_u4
and then adding to the end of the file the details of the new binary:
nc_q4_c1_u2_u4_LDFLAGS = $(AM_LDFLAGS)
nc_q4_c1_u2_u4_CXXFLAGS = $(AM_CXXFLAGS) \
-D_GLIBCXX_USE_NANOSLEEP \
-D_GLIBCXX_USE_SCHED_YIELD \
-DLIST_DIMENSION_NAMES=q4,c1 \
-DLIST_VARIABLE_TYPES=u2,u4 \
-DVERSION=\"$(VERSION)\"
nc_q4_c1_u2_u4_SOURCES =\
$(nc_SOURCES)
Then you should build and install again the nanocube binaries.